By Kevin E. Noonan —
 The domestic dog (Canis lupus familiaris) has been the subject of many genetic studies, particularly since the dawning of the age of Whole Genome Sequencing in the late 20th Century. These studies have elucidated some interesting facts about "man's best friend," some of which have been discussed here (see "The Genetic Basis of Coat Variation in Dogs"; "Leg Length Variation in Dogs and its Relevance to Human Mutations"; "From Toy Poodle to Rottweiler: Why Is Fido So Small (or Large)?"; "Selection for Facial Features in Domestic Dogs: The Evolution of Cuteness") and comparators to related species (see "Red Fox Genome Sheds Light on Domesticated Dogs (and Maybe Humans)").
The domestic dog (Canis lupus familiaris) has been the subject of many genetic studies, particularly since the dawning of the age of Whole Genome Sequencing in the late 20th Century. These studies have elucidated some interesting facts about "man's best friend," some of which have been discussed here (see "The Genetic Basis of Coat Variation in Dogs"; "Leg Length Variation in Dogs and its Relevance to Human Mutations"; "From Toy Poodle to Rottweiler: Why Is Fido So Small (or Large)?"; "Selection for Facial Features in Domestic Dogs: The Evolution of Cuteness") and comparators to related species (see "Red Fox Genome Sheds Light on Domesticated Dogs (and Maybe Humans)").
But as with other organisms, a great deal of the genetic sequence set forth early in the 21st Century was incomplete due to technical limitations. Chief amongst these was difficulty in obtaining reliable sequence information on sequences repeated in many places in the genome, which interfered with reliable assembly of sequenced portions (termed "contigs") into longer (ideally, chromosomal-length) linear sequence assemblies (termed "scaffolds). Improvements in sequencing technology now permit better sequence determinations for high GC and highly repetitive regions.
The existence of a prior reference genome (produced from a female boxer named Tasha), combined with these new sequencing tools, was used by an international team of researchers* in a paper published in March of this year in the Proceedings of the National Academy of Sciences (USA) entitled "Long-read assembly of a Great Dane genome highlights the contribution of GC-rich sequence and mobile elements to canine genomes." These scientists reported elucidation of the genomic sequence from a female Great Dane named Zoey, wherein they were able to identify and eliminate (in large part) incorporation of false contigs due to repetitive ends and fragment ends that map to multimer genomic locations. Using alignment with the reference dog genome revealed gaps between contigs as earlier reported in dog genomic DNA. They report finding 373 additional sequence regions spanning contigs having N50 of 30kb and a total length of 10.5 Mbp. The resulting aligned scaffolds were assigned to dog chromosomes to constitute a genomic sequence for the dog genomic complement of 78 chromosomes.
From this sequence the researchers were able to identify 22,182 protein-coding gene models; full-length matches with the reference canine genome were found for only 84.9% (18,834) of all protein-coding gene models, and almost full-length alignments were found for 93% (20,670) of the models; in addition they identified 49 protein-coding genes not present in the earlier canine reference genome. Also annotated were 7,049 long noncoding RNAs that included 84 with no or only partial alignment to the reference canine genome. From this comparison they further were able to appreciate that the assembly from the Great Dane genome spanned the majority of sequence gaps not having been sequenced in the reference dog genome. Mapping of these gaps showed that 2,151 gaps (16.8% of gaps) overlapped the transcription start site of a predicted protein-coding gene and were found to be extremely GC rich (67.3%). Further, a subset of the resolved gaps, having a median 80.95% GC content, were found to be preferentially localized (i.e., at a frequency greater than random chance) at transcription start sites and recombination hotspots. About 12% (1,457 of 12,304 on the autosomes) of gap segments were found to be located within 1 kbp of a hotspot (the expected percentage of such location was closer to 3%). These hotspot-adjacent segments were extremely GC rich; 5,553 such identified segments had a GC content greater than that what would be expected from random sequence permutations. The total extent of these extreme GC segments spanned 4.03 Mbp in the Great Dane sequence assembly, were found to have "a median length of 531 bp, a median GC content of 80.95%, and are located much closer to transcription start sites (median distance of 290 bp) and recombination hotspots (median distance of 68.7 kbp) than expected by chance."
Analysis of the Great Dane genome assembly with the reference canine genome identified 16,834 deletions (median size: 207 bp) and 15,621 insertions (median size: 204 bp) in Great Dane DNA. Genetic assessment of these sequences revealed predominantly the presence of two forms of "retrotransposon insertion/deletion polymorphisms" which included dimorphic canine short interspersed elements (SINECs) (16,221 copies) and dimorphic long interspersed element-1 sequences (LINE-1_Cfs) (1,121 copies). The 3' flanking sequence for the LINE-_Cfs elements suggested "multiple retrotransposition-competent LINE-1_Cfs segregate among dog populations."
The researchers further reported a length distribution of the detected variants having "a striking bimodal pattern, with clear peaks at ∼200 bp and ∼6 kbp, consistent with the size of SINEC and LINE-1 sequences," as shown in this Figure:
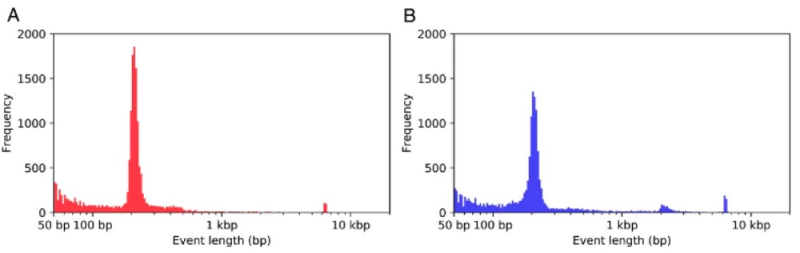
The location of these sequences was associated with insertions and deletions, the researchers reporting that inspection of the sequences in the 150-250-bp range (dimorphic SINEC elements) were found at 7,298 deletion and 6,071 insertion sites, and that "LINE-1 sequences accounted for 339 deletions and 581 insertions longer than 1 kbp." When combined with data from the reference canine genome there were at least 16,221 dimorphic SINEC and 1,121 dimorphic LINE-1 sequences identified. "Hallmarks of retrotransposition" were identified at these sites, wherein the SINEC and LINE-1 sequences were flanked by "target site duplications having a 15 bp median size with the elements ending in poly(A) tracts having median lengths of 9 bp to 12 bp."
To test whether these sequences were capable of retrotransposition, a particular sequence was cloned that had intact open reading frames encoding the ORF1p and ORF2p predicted proteins and lacked mutations expected to disrupt protein function. This element was introduced into human cells in vitro and shown to be capable of retrotransposition, as illustrated in this Figure:
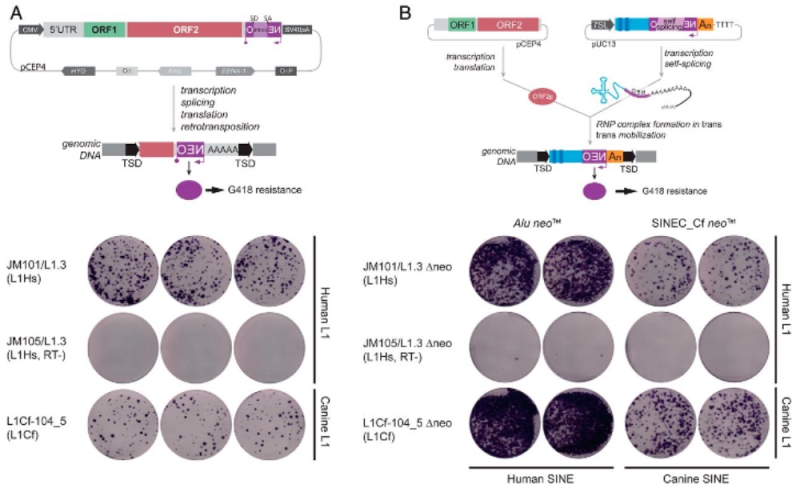
This element also capable of mobilizing both the canine SINEC elements and analogous human Alu elements. The researchers speculated that this result was consistent with ongoing retrotransposon activity as a driver of canine genetic variation.
The researchers set forth a synopsis of their results as follows. They had identified 49 predicted protein-coding genes from the Great Dane assembly that were not found in the canine genome, as well as 2,151 protein-coding gene models having a transcription start position located in a gap in the reference genome sequence. The existence of high GC-content sequences in canine promoter regions distinguishes the preferential location of recombination events in dogs, which lack a functional PRDM9 gene known to mediate recombination in other mammals; see Paigen & Petkov, 2018, "PRDM9 and its role in genetic recombination," Trends Genet. 34, 291–300, and Auton et al., 2013, "Genetic recombination is targeted towards gene promoter regions in dogs," PLoS Genet. 9, e1003984. The researchers assert that "[t]he presence of extremely GC-rich segments likely reflects a key aspect of canine genome biology" as a consequence of these findings. Further, a comparison of the number of single nucleotide variants (i.e., differences) found in Zoey and Tasha (3.57 million) was lower than found in similar comparison between humans (4.1-5.0 million). In contrast, the levels of LINE-1 and SINEC dimorphism between these two dog genomes was "disproportionately large," there being "an ∼17-fold increase in SINE differences (16,221/915) and an eightfold increase in LINE differences (1,121/128) compared to the numbers found among humans" (indeed, the researchers report that "more dimorphic SINEs were found between these two breed dogs than have been found in studies of thousands of humans"). They note that these results are consistent with prior studies, including Wang & Kirkness, 2005, "Short interspersed elements (SINEs) are a major source of canine genomic diversity," Genome Res. 15, 1798–808.
The authors conclude by saying:
[O]ur study suggests that retrotransposition is an ongoing process that continues to affect the canine genome. We provide proof-of-principle evidence that dog genomes contain LINE-1 and SINEC elements that are capable of retrotransposition in a cultured cell assay. We also identified two LINE-1 lineages with the same 3′ transduced sequence associated with multiple elements, suggesting the presence of multiple canine LINE-1s that are capable of spawning new insertions. Additionally, analysis of 3′ transduction patterns suggests the presence of additional active LINE-1s in canines that have yet to be characterized. Thus, a full understanding of canine evolution and phenotypic differences requires consideration of these important drivers of genome diversity.
* From the Department of Biological Sciences, Bowling Green State University; the Department of Human Genetics, Department of Computational Medicine and Bioinformatics, and the Department of Internal Medicine, University of Michigan; the Université Côte d'Azur, CNRS, INSERM, Institut de Recherche sur le Cancer et le Vieillissement de Nice, Nice; the Université de Rennes 1, CNRS, Institut de Génétique et Développement de Rennes−UMR 6290, Rennes; and the Department of Biomedical Sciences, Cornell University, Ithaca, NY 14850.

Leave a comment