By Kevin E. Noonan —
 Hiram Bentley Glass and classical geneticists of the Twentieth Century elucidated some of the ways that genetics could inform regarding human populations and their history, using observations like genetic drift (famously, among the Amish) and the "founder effect." The "genomics" revolution of the last thirty years has extended these observations, for human populations as well as many other animal and plant species (see, e.g., "Genetic Assessment of Squash Genomes in Related Species"; "The Domestication History of Apples Revealed by Genomic Analysis"; and "Domestic Cat Genome Sequenced"). Recently, several outstanding questions regarding the genetics of sweet potato were resolved, in a report in Current Biology, entitled "Reconciling Conflicting Phylogenies in the Origin of Sweet Potato and Dispersal to Polynesia," resulting from genomic and chloroplast DNA analyses that establish the phylogenetic, temporal and geographical relationships between this important crop species and its wild naturally occurring relatives.
Hiram Bentley Glass and classical geneticists of the Twentieth Century elucidated some of the ways that genetics could inform regarding human populations and their history, using observations like genetic drift (famously, among the Amish) and the "founder effect." The "genomics" revolution of the last thirty years has extended these observations, for human populations as well as many other animal and plant species (see, e.g., "Genetic Assessment of Squash Genomes in Related Species"; "The Domestication History of Apples Revealed by Genomic Analysis"; and "Domestic Cat Genome Sequenced"). Recently, several outstanding questions regarding the genetics of sweet potato were resolved, in a report in Current Biology, entitled "Reconciling Conflicting Phylogenies in the Origin of Sweet Potato and Dispersal to Polynesia," resulting from genomic and chloroplast DNA analyses that establish the phylogenetic, temporal and geographical relationships between this important crop species and its wild naturally occurring relatives.
The paper, from an international group of researchers* from the University of Oxford, Oregon State University, International Potato Center (Lima, Peru) and Duke University, used genome skimming and target DNA capture on 199 specimens comprising sweet potato (Ipomoea batatas) and all its wild relatives (I. trifida, I. triloba, I. ramosissima, I. cordatotriloba, and I. leucantha). One of the most consumed crops worldwide, the species originated in the Caribbean region (Central and northeastern South America and the islands comprising the region) but is also endemic to Polynesia. In addition to the evolutionary relationship between I. batatas and its wild relatives, this geographical distribution pattern has long raised the question of whether dispersion occurred without human intervention (wind, water, birds) or whether there were humans transiting the Pacific much earlier than anthropological evidence suggests. The genetic evidence shows that human intervention is not necessary to explain sweet potato presence in Polynesia based on comparisons of the relationship (and divergence) of the Polynesian species (Ipomoea littoralis Blume) with I. batatas and other New World members of its family.
The researchers report that their dataset consisted of 199 individuals representing all sixteen species in Ipomoea series Batatas and twenty-four other species across the Ipomoea genus. These included fresh samples growing contemporaneously as well as herbarium specimens dating back to 1769. Probes for 605 putative single copy nuclear regions of Ipomoea were derived from a comparison between genomic information from I. lacunosa and coding sequence-restricted data from the related Solanum tuberosum (potato). The team interrogated genomic regions with a one-to-one match at 70% identity along at least half the length of a Solanum CDS and the results were filtered to retain Ipomoea loci that were at least 1000 bp long. From these loci 100 bp-long RNA probes were developed, excluding probes with GC content < 25%. In addition, the researchers obtained complete chloroplast DNA sequences from each specimen.
From these comparisons the scientists concluded that the I. batatas sweet potato species is most closely related to I. trifida. Speciation arose from a first autohexaploidy event, with the related species having the following family tree set forth in A:
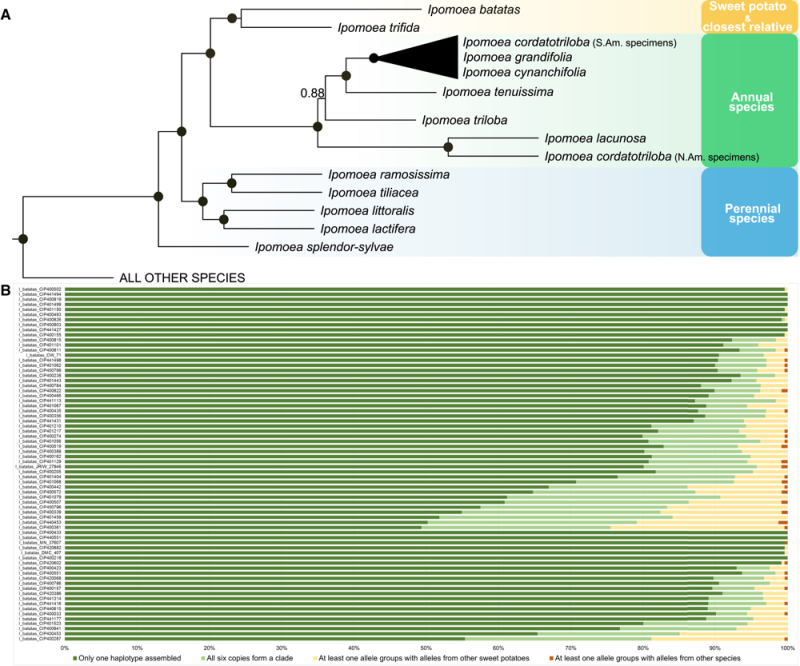
Muñoz-Rodríguez et al., 2018, Current Biology 28(8): 1246-56, Figure 2
Sweet potato is also the only member of this extended family whose evolution contains an autohexaploidy event. This origin was supported by an analysis of the six alleles in each I. batatas genome, which the researchers found were more closely related to each other than to any other species (shown in B).
When did I. batatas diverge from I. trifida? Using nuclear DNA comparisons over 21 nuclear regions, the researchers estimated that divergence occurred at least 800,000 years ago; there was also evidence of a population bottleneck ~640,000 years ago.
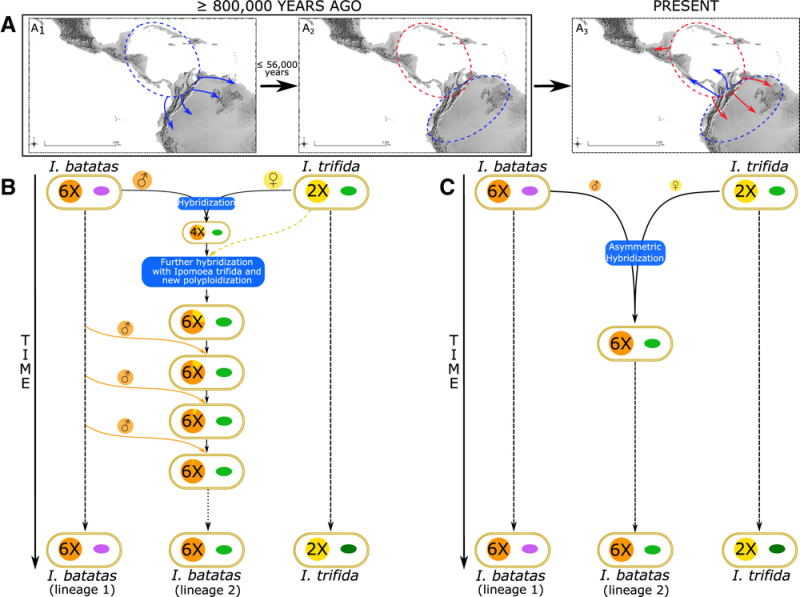
Whole chloroplast genome analysis was performed and revealed two genetically distinct lineages, with one (termed CL2) being more closely related to chloroplasts contained in I. trifida. The relationship between the two chloroplast species compared with chloroplasts from I. trifida reported showed no shared insertion/deletion events (indels) between CL1 and CL2, but that I. batatas CL2 chloroplasts shared indels in common with I. trifida chloroplasts. The results of these analyses suggested that the sweet potato species had undergone two speciation events: the first, autohexaploidy from an I. trifida-like ancestor, followed by later introgresssion (occurring within about 56,000 years from the autohexapolidy event) by I. trifida, resulting in two populations of chloroplasts in natural species. The Figure represents this history:
Muñoz-Rodríguez et al., 2018, Current Biology 28(8): 1246-56, Figure 4
This Figure shows two alternative histories to explain the two chloroplast lineages in natural populations, with the evidence being more consistent with Figure C, resulting in an I. batatas species having CL1 and Cl2 chloroplasts.
Turning to the genetic evidence for Polynesian dispersion (which extends today from Polynesia to Madagascar), comparison between the Ipomoea littoralis Blume species and I. batatas indicated (through genetic differences that would have been familiar to Bentley Glass and colleagues) that the sweet potato dispersed to Polynesia 111,500 – 139,000 years ago, predating human contact and putting to rest speculation for the occurrence of trans-Pacific exploration by Polynesia populations earlier than anthropological and other more conventional data suggest.
One of the benefits noted by the scientists in their paper is the practical consequence that, by knowing the relationships between sweet potato and its relatives it may be possible to improve production or other aspects of breeding for a commercially important crop. More broadly, though, researches such as these extend to more quotidian questions the reach of modern genetic analysis to address long-standing historical conundrums (both human and natural) that were hoped for by the pioneers of the genomics revolution. Indeed, in view of the transitory nature of history and its artifacts, it is likely that many such questions can only reliably be answered from the history of life written in the genetic code of the organisms that have survived (which makes preserving them all the more important).
*Pablo Muñoz-Rodríguez, Tom Carruthers, John R.I. Wood, Bethany R.M. Williams, Kevin Weitemier, Brent Kronmiller, David Ellis, Noelle L. Anglin, Lucas Longway, Stephen A. Harris, Mark D. Rausher, Steven Kelly, Aaron Liston, and Robert W. Scotland
Image of Ipomoea batatas, Sweet Potato by Llez, from the Wikimedia Commons under the Creative Commons Attribution-Share Alike 3.0 Unported license.

Leave a comment