By Kevin E. Noonan —
 On December 3rd, Senior Party Sigma-Aldrich filed its Preliminary Motion No. 1 that asked the Board to deny the Broad Institute, Harvard University, and MIT (collectively, Broad) benefit of its U.S. Provisional Application No. 61/736,527, filed December 12, 2012 (termed "P1"), pursuant to 37 C.F.R. § 41.121(a)(1) and S.O. ¶¶ 121 and 208.4.2. On March 16th, Broad filed its Opposition, and on April 26th, Sigma-Aldrich filed its Reply.
On December 3rd, Senior Party Sigma-Aldrich filed its Preliminary Motion No. 1 that asked the Board to deny the Broad Institute, Harvard University, and MIT (collectively, Broad) benefit of its U.S. Provisional Application No. 61/736,527, filed December 12, 2012 (termed "P1"), pursuant to 37 C.F.R. § 41.121(a)(1) and S.O. ¶¶ 121 and 208.4.2. On March 16th, Broad filed its Opposition, and on April 26th, Sigma-Aldrich filed its Reply.
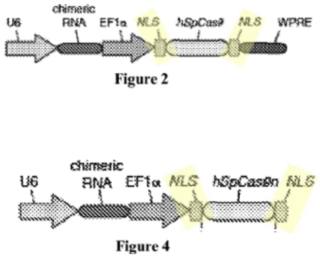
The basis for Sigma-Aldrich's Motion was that Broad's P1 application did not disclose a constructive reduction to practice of an embodiment within the scope of the Count of the interference as declared. Sigma-Aldrich specifically contended that the P1 application does not show that Broad's inventors had achieved a CRISPR-Cas9 system that cleaved a targeted DNA molecule and altered the expression of the gene product of that cleaved molecule. Sigma-Aldrich argued that Broad merely identified the existence of CRISPR-mediated DNA cleavage in a eukaryotic cell and did not establish changes in gene expression as a result of such cleavage. Sigma-Aldrich further argued that Broad's P1 application did not disclose introducing Cas9 protein into a eukaryotic cell with only one linked nuclear localization signal ("NLS") that resulted in CRISPR-mediated gene editing, Sigma-Aldrich asserting that all of the P1 disclosure involved Cas9 modified with 2 NLS sequences, as illustrated by Figures from that application:

In its Opposition, Broad first asked the Board to dismiss Sigma-Aldrich's Motion for being moot should the Board grant Broad’s Motion No 1 to change the Count. On the merits, Broad argued that it was within the skill of the ordinary artisan to alter gene expression in eukaryotic cells in light of prior art methods, specifically zinc-finger nucleases ("ZFNs") and transcription activator-like effector nucleases ("TALENs"). The distinction that CRISPR provided was that it is a system that could be "easily and quickly designed, in practice, to specifically modify any sequence of any genome; having more than one technology available will help achieve this goal" (emphasis added), and that once its inventors had reduced eukaryotic cell CRISPR to practice they provided the capacity to alter gene expression as well. Broad also argued that the Board should grant Sigma-Aldrich's Motion No. 1 only if it establishes that P1 does not show possession of either half of the Count of this interference as declared, pursuant to 37 C.F.R. §§ 41.121(b), 41.208(b); S.O. ¶ 121.3; Bd.R. 201; and S.O. ¶ 208.4.2. But of course Broad argued the Board should not do so, because (in a recurrent theme) P1 disclosed "a eukaryotic CRISPR-Cas9 system capable of programmable, generalizable, site-specific DNA cleavage, including for both donor template integration (which can be used for inserting sequences into the DNA) and multiplexing (introducing multiple concurrent breaks in DNA to cause large deletions)." But Broad's definition of the term "altering gene expression" included altering the sequence of the gene, whereas Sigma-Aldrich contended that the term meant altering gene expression levels. Broad asserted that the term "altering gene expression" should be not limited to changing gene expression levels, which Broad contends is "an overly narrow and erroneous understanding of what altering expression means" under a broadest reasonable interpretation standard. Finally, Broad contended that P1 disclosed embodiments falling within the "other half" of the McKelvey Count, relating to Cas9 specific having a single nuclear localization sequences (NLS).
In its Reply, Sigma-Aldrich begins by stating that Broad has not refuted its argument that the P1 application is limited to disclosing altering the sequence of a target gene, and that Broad cited to no disclosure expressly relating to altering gene expression (making this definitional dispute a focus for the Board's consideration). Sigma-Aldrich asserts Broad's is an inherent disclosure position, and that there is no basis for the Board to consider altering a target gene sequence to be equivalent to altering gene expression (except for the fact that altering a gene sequence could be considered to reduce gene expression of the unaltered gene to zero and creating de novo expression of the sequence-altered gene). Sigma-Aldrich characterizes the Broad arguments in this regard as "hand-waiving and conclusory assumptions." Sigma-Aldrich cites Broad's arguments in earlier Interferences Nos. 105,048 and 106,115 "acknowledging and affirmatively arguing" that "altering gene expression" is an affirmative limitation and, in any event, the phrase is limiting because it imparts a further functional requirement on the Count, illustrating this point colorfully using Broad’s claims as an example:

The brief further illustrates the effect of this limitation graphically:
Further, Sigma-Aldrich notes that Broad argued in the '115 Interference that P1 disclosed CRISPR cleavage and not altered gene expression, raising an inconsistency in Broad's position if not an explicit estoppel (in light of the fact that Broad has also taken the position that the Board should rely on its Decision on Priority in Broad’s favor in the '115 Interference in this interference. Also in this regard Sigma-Aldrich argues that the distinction it is drawing in this interference between altering gene sequence and gene expression is one Broad made in the P1 application. Sigma-Aldrich then turns to prosecution history of Broad patents, in particular U.S. Patent No 8,697,359, to support the distinction it is drawing between altering gene sequence and altering gene expression, reproducing one such argument and supporting graphical data:
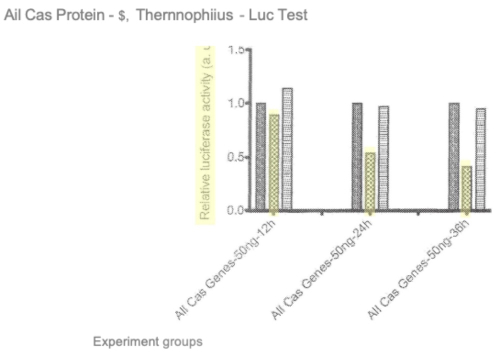
illustrating a reduction in luciferase activity. Sigma-Aldrich also argues that the plain and ordinary meaning of the phase "altering gene expression" means just that (changing gene expression levels) and does not encompass merely altering a sequence. Sigma-Aldrich follows these arguments with others, amounting in total over a dozen instances where the term "altering gene expression levels" makes the distinction is argues here with altering gene sequence. While acknowledging that Broad's patents and applications contain conventional "boilerplate" disclosure comprising "laundry lists" of methods for detecting alterations in gene expression levels Sigma-Aldrich asserts that "Broad P1 nowhere conducts any of those well-known and conventional analyses to evaluate gene product expression" and "never even mentions altering expression of a gene product."
Turning to the NLS element of the Count, Sigma-Aldrich argues that Broad is asserting "fictitious embodiments" and that Broad’s attempt to "mix and match" different embodiments between one- and two-NLS embodiments do not anticipate Sigma-Aldrich's Claim 31 (noting that Broad criticized CVC for making similar arguments in the '115 Interference) and that "a single NLA does not anticipate [constructs comprising] 2 NLSs."

Leave a comment