By Kevin E. Noonan —
 On March 16th, Junior Party the Broad Institute, Harvard University, and MIT (collectively, Broad) filed its Opposition to Substantive Preliminary Motion No. 1 filed on December 3rd by Senior Party Sigma-Aldrich.
On March 16th, Junior Party the Broad Institute, Harvard University, and MIT (collectively, Broad) filed its Opposition to Substantive Preliminary Motion No. 1 filed on December 3rd by Senior Party Sigma-Aldrich.
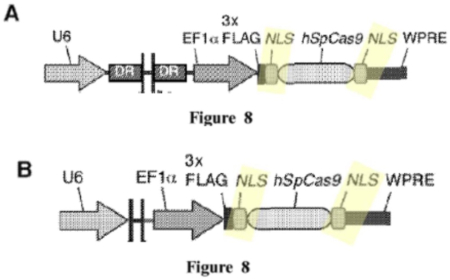
To recap, Sigma-Aldrich's Substantive Preliminary Motion No. 1 asked the Board to deny Broad benefit of its U.S. Provisional Application No. 61/736,527, filed December 12, 2012 (termed "P1"), pursuant to 37 C.F.R. § 41.121(a)(1) and S.O. ¶¶ 121 and 208.4.2. The basis for Sigma-Aldrich's motion was that this application does not disclose a constructive reduction to practice of an embodiment within the scope of the Count of the interference as declared. Sigma-Aldrich specifically contended that the P1 application does not show that Broad's inventors had achieved a CRISPR-Cas9 system that cleaved a targeted DNA molecule and altered the expression of the gene product of that cleaved molecule. Sigma-Aldrich argued that Broad merely identified the existence of CRISPR-mediated DNA cleavage in a eukaryotic cell and did not establish changes in gene expression as a result of such cleavage. Sigma-Aldrich further argued that Broad's P1 application did not disclose introducing Cas9 protein into a eukaryotic cell with only one linked nuclear localization signal ("NLS") that resulted in CRISPR-mediated gene editing, Sigma-Aldrich asserting that all of the P1 disclosure involved Cas9 modified with 2 NLS sequences, as illustrated by Figures from that application:
Broad's Opposition begins by asking the Board to dismiss Sigma-Aldrich's Motion No. 1 as moot in the event that the Board grants Broad's Motion No 1 to change the Count. Barring that, Broad contends that the prior art's facility for altering gene expression in eukaryotic cells with zinc-finger nucleases ("ZFNs") and transcription activator-like effector nucleases ("TALENs") rendered it within the ordinary skill in the art to alter gene expression in eukaryotic cells using CRIPSR (although this argument seems inconsistent with Broad's arguments regarding the routine nature vel non of genetic manipulation in eukaryotic cells made elsewhere). Broad argues that CRISPR provided something those earlier methods had not, a system that could be "easily and quickly designed, in practice, to specifically modify any sequence of any genome; having more than one technology available will help achieve this goal" (emphasis added). Broad's argument is that its inventors' achievement of CRISPR-mediated cleavage in eukaryotic cells was enough to provide the skilled artisan with the ability to modify gene expression as well. Broad contends that these assertions are supported by disclosure in P1 as well as its manuscript submitted on October 5, 2012, and statements made by the manuscript's reviewers. In these arguments, Broad asserts that disclosure of CRISPR for "mammalian genome engineering" would be understood by the skilled worker to include gene expression alterations. Broad further asserts that Sigma-Aldrich's position is inconsistent with positions taken by CVC and ToolGen in their interferences against Broad (which is more properly understood to be the case because those parties did not pursue expressly this aspect of CRISPR technology in eukaryotic cells rather than being a concession about what Broad's P1 application disclosed).
Broad argues that the Board should grant Sigma-Aldrich's Motion No. 1 only if it establishes that P1 does not show possession of either half of the Count of this interference as declared, pursuant to 37 C.F.R. §§ 41.121(b), 41.208(b); S.O. ¶ 121.3; Bd.R. 201; and S.O. ¶ 208.4.2, as well as the holding in Kubota v. Shibuya, 999 F.2d 517, 521 (Fed. Cir. 1993). According to Broad, Sigma-Aldrich fails because its possession of an invention falling within the scope of the Count requires P1 to disclose "a eukaryotic CRISPR-Cas9 system capable of programmable, generalizable, site-specific DNA cleavage (this phrase, italicized here, is repeated like a mantra throughout the brief), including for both donor template integration (which can be used for inserting sequences into the DNA) and multiplexing (introducing multiple concurrent breaks in DNA to cause large deletions)" and that Sigma-Aldrich does not challenge possession of this invention. Broad points to over a dozen instances in the P1 specification that it contends disclose that its CRISPR-Cas9 invention can alter gene expression. Broad also argues that the term "altering expression" should be construed broadly to include altering the sequence of the gene as well as altering gene expression levels (the latter meaning being the one Sigma-Aldrich contends is the correct one here). The focus of Broad's argument is that the skilled worker would understand its disclosure of "a programmable, generalizable system that would induce site-specific cleavage of DNA in a eukaryotic cell" would show possession of embodiments that alter expression of specific eukaryotic genes without demonstration or even express disclosure of such embodiments based, in part, on "prior work with systems such as ZFNs and TALENs."
Broad's arguments in this regard present something of an inverse déjà vu for anyone following the CRISPR interference saga. Broad argues that Sigma is wrong in its "flawed premise that a POSA would need working examples with assays showing measurements of the level of altered gene expression, e.g., altered levels of protein expression, to find possession" (emphasis in brief) and that based on the "inherent complexities and unpredictability of the eukaryotic processes that impact gene expression," a POSA would not have found possession, "particularly without any demonstrated showing of DNA cleavage resulting in alteration of the targeted gene's expression." The skilled worker would know how to adapt CRISPR for these purposes, Broad now argues in its Opposition, whether P1 discloses "working examples of a specific assay or not." Broad asserts in support of this argument the comments of "impartial reviewers" of its scientific manuscript submitted October 5, 2012 (which, to be fair, is a document that is not identical to P1 and is not subject to the same standards of disclosure as P1). Broad further asserts what is expressly disclosed in P1 (creation of insertion/deletion gene alterations using CRISPR, as well as specific insertions and deletion) in support of its argument for possession. Broad argues that the P1 disclosure contains working examples showing altered gene expression (while stating in the same rhetorical breath that it is unnecessary for showing possession). Such examples are the same examples cited above (indels, insertions, deletions) which, one supposes, would alter gene expression with regard to what sequences are expressed (provided the target is transcriptionally active) rather than the levels at which they are expressed.
Broad further asserts that Sigma-Aldrich failed to address whether the phrase "whereby expression of at least one gene product is altered" is an affirmative limitation (because inter alia a Board decision that it is not would decide the matter in Broad's favor). And Broad asserts as an independent basis for opposition that the term "altering gene expression" should be not limited to changing gene expression levels, which Broad contends is "an overly narrow and erroneous understanding of what altering expression means" under a broadest reasonable interpretation standard (an argument that could have traction with the Board), citing a portion of Broad's P1 specification that imperfectly supports this construction (while strenuously arguing that Sigma-Aldrich's interpretation of other portions of the P1 specification that seems to support its position would not be so understood by the skilled worker).
Broad also challenges Sigma-Aldrich's contention that P1 does not show possession of the "other" half of the Count, which requires a Cas9 protein having a single NLS sequence. Broad argues that "a working example with one NLS was not necessary once Zhang B1 disclosed: (1) a functional eukaryotic CRISPR-Cas9 system to accomplish donor template integration, and (2) the use of a Cas9 with "one or more nuclear localization sequences" with disclosure of numerous embodiments in which the Cas9 "comprises one or more nuclear localization sequences of sufficient strength to drive accumulation of said CRISPR enzyme in a detectable amount in the nucleus of a eukaryotic cell." Broad relies on deposition testimony from its expert, Dr. Seeger, that the disclosure in Figure 1B "showed that the system functioned in the eukaryotic cells with both one and two NLS" and further language in the P1 specification that CRISPR-Cas9 complexes provided by the invention comprise Cas9 protein comprising "one or more nuclear localization sequences." Further, Broad contends, "the "'written description requirement does not demand either examples or an actual reduction to practice' in every case," citing Ariad Pharms., Inc. v. Eli Lilly and Co., 598 F.3d 1336, 1352 (Fed. Cir. 2010). Broad asserts that, despite disclosure of nothing more than nuclear localization in P1 Sigma-Aldrich has not satisfied its burden of showing that a working example of a Cas9 construct comprising a single NLS was necessary and thus that the Board should deny Sigma-Aldrich's motion with regard to the single NLS limitation portion of the Count.

Leave a comment