By Kevin E. Noonan —
 The portion of the eukaryotic world inhabited by plants exhibits a genetic complexity not shared by members of the animal world (see, for example, "Rose Genome Reveals Its Exquisite Complexities"). The strawberry (Fragaria × ananassa) genome, recently explicated in a paper published last month in Nature Genetics, provides yet another example of this characteristic.
The portion of the eukaryotic world inhabited by plants exhibits a genetic complexity not shared by members of the animal world (see, for example, "Rose Genome Reveals Its Exquisite Complexities"). The strawberry (Fragaria × ananassa) genome, recently explicated in a paper published last month in Nature Genetics, provides yet another example of this characteristic.
The paper, entitled "Origin and evolution of the octoploid strawberry genome," which was authored* by scientists from several academic institutions and genomics companies, provides a complete genomic sequence for the allo-octoploid ("2n = 8X = 56" chromosomes) strawberry genome, which is appreciated as being "an interspecific hybrid between wild octoploid progenitor species approximately 300 years before present." The immediate progenitor species, Fragaria virginiana and Fragaria chiloensis, are themselves products of fusion from four earlier progenitor species about one million years ago. The strawberry is a member of the Rosaceae family, which includes almonds, apples, peaches, and roses, in addition to the strawberry. In the wild, there are 22 known species of strawberry, ranging in genome size from diploid (2n = 2x = 14) to decaploid (2n = 10x = 70). As explained in the paper, "[a]llopolyploids face the challenge of organizing distinct parental subgenomes—each with a unique genetic and epigenetic makeup shaped by independent evolutionary histories—residing within a single nucleus." An advantage of analyzing the strawberry genome is that this species has maintained a complete set of "homeologous" chromosomes from all four parental subgenomes and has thus avoided the "mixing" (presumably through recombination) of stretches of genome from different parental chromosomes found, inter alia, in the rose genome. The existence of gene sequences from "extant relatives" of the octoploid strawberry facilitate assignment of homeologs to each parental subgenome. The strawberry genome comprises 813.4 megabases (Mb) and has a high heterozygosity as well as ploidy level.
This genome assembly was prepared from a particular Fragaria × ananassa cultivar 'Camarosa', at least in part because this is one of the most widely cultivated strawberry worldwide. The total length of the sequenced assembly is 805,488,706 bp distributed among 28 "pseudochromosomes." This sequence contained "108,087 protein-coding genes along with 30,703 genes encoding long noncoding RNAs (lncRNAs), which were subdivided into 15,621 long intergenic noncoding RNAs, 9,265 antisense overlapping transcripts (AOT-lncRNAs), and 5,817 sense overlapping transcripts (SOT-lncRNAs)." Most of a collection of 1,440 "core genes" in common between land plants were identified in this assembly. 36% of the sequenced genome comprised transposable elements, including "DNA transposons, long-terminal-repeat retrotransposons (LTR-RTs; for example, Copia and Gypsy), and non-LTR retrotransposons," with LTR-RTs being the most abundant class (28%).
The authors used this information to identify the existing diploid relatives (and hence progenitors) of each of the four subgenomes that make up the octoploid strawberry genome. This analysis was based on an assembly of 31 "transcriptomes," i.e., the transcribed portion of the genomes, from every described diploid Fragaria species and comparison of 19,302 nuclear genes. These results confirmed the participation of two previously identified diploid progenitor species, F. iinumae and Fragaria nipponica (both of which are endemic to Japan) and revealed two strong candidates for the previously unknown progenitor species as F. vesca subsp. Bracheata (endemic to the western part of North America, spanning Mexico to British Columbia) and Fragaria viridis (geographically distributed in Europe and Asia. The results of this reconstruction supported a North American origin of the modern allo-octoploid strawberry. These relationships are illustrated in Supplementary Fig. 7:
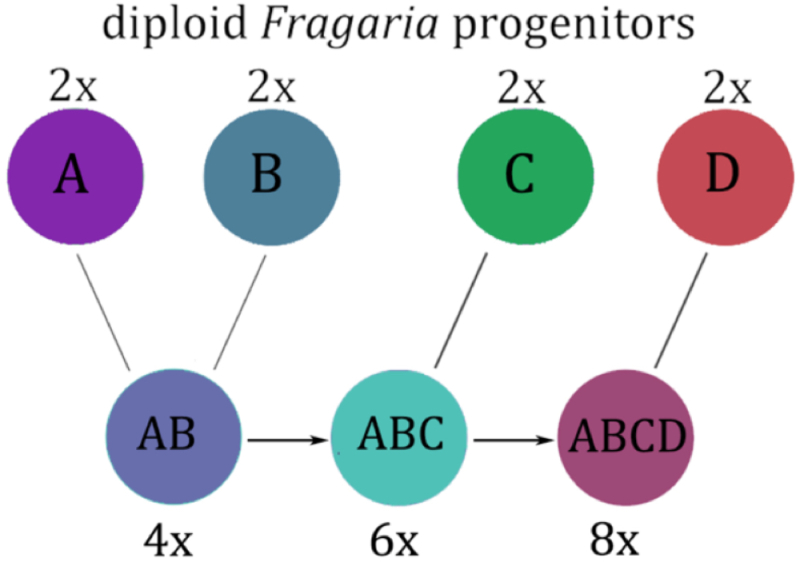
where Species A and B represent F. nipponica (purple) and F. iinumae (blue), respectively, to form a tetraploid. The ancestral tetraploid strawberry hybridized with F. viridis (species C, green) to form the ancestral hexaploid species, which hybridized with F. vesca ssp.bracteata (species D, red) to form the octoploid strawberry.
In many polyploidy plant species there develops through evolution a "dominant" subgenome from one of the progenitor species. The authors used the genomic sequencing results to identify a dominant subgenome that was contributed by the F. vesca progenitor, which "has retained 20.2% more protein-coding genes and 14.2% more lncRNA genes, and has overall 19.5% fewer TEs than the other homoeologous chromosomes." This subgenome also exhibits about 41% more tandem gene duplications (both "a greater number of tandem gene arrays as well as larger average tandem-gene-array sizes on six of seven homoeologous chromosomes"). A possible explanation/consequence of this dominance is a bias for disease-resistance genes in the F. vesca progenitor-derived subgenome. These genes included 423 nucleotide-binding-site leucine-rich-repeat genes (NBS-LRRs), which included 79 encoding toll interleukin receptor and 24 encoding genes involved in resistance to powdery mildew. This subgenome also contained 20 genes for R proteins that "recognize pathogen effectors through integrated decoy domains," consistent with the strawberry genome containing "a greatly expanded set of 105 diverse domains that are fused to the R-protein structures and have the potential to function as integrated decoys."
While the "pseudochromosomes" of the strawberry genome have remained relatively more intact than in other species, these authors report detecting homeologous exchange (HE) sites as well as instances of gene conversion, these also biased (7.3-fold) in favor of the F. vesca progenitor. These regions range in size from single genes to megabase-sized chromosomal regions (which the authors note have been seen in other alloploidy species including cotton and bread wheat). Further analysis showed a transcriptome-wide expression dominance of genes derived from the F. vesca progenitor, which they hypothesize results "n certain biological pathways being largely controlled by a single dominant subgenome,' including those involved in strawberry flavor, color, and aroma. Evidence for this hypothesis include that "F. vesca homoeologs in octoploid strawberry are responsible for 88.8% of the biosynthesis of anthocyanins, the metabolites responsible for the red pigments in ripening strawberry fruit; 89.2% of the biosynthesis of geranyl acetate, a terpene associated with fruit aroma; and 95.3% of the biosynthesis of fructose associated with sweetness."
The authors provide their own assessment of the significance of their strawberry genome assembly:
This reference genome should serve as a powerful platform for breeders to develop homoeolog-specific markers to track and leverage allelic diversity at target loci. Thus, we anticipate that this new reference genome, combined with insights into subgenome dominance, will greatly accelerate molecular breeding efforts in the cultivated garden strawberry.
 North-polar projection of present day. Geographic distributions of extant relatives of the diploid (2×) progenitors of Fragaria × ananassa, the putative intermediate tetraploid (4×) and hexaploid (6×) progenitors of Fragaria × ananassa, and extant wild octoploid (8×) species in North America.
North-polar projection of present day. Geographic distributions of extant relatives of the diploid (2×) progenitors of Fragaria × ananassa, the putative intermediate tetraploid (4×) and hexaploid (6×) progenitors of Fragaria × ananassa, and extant wild octoploid (8×) species in North America.
* Patrick P. Edger**, Robert VanBuren, Marivi Colle, Ching Man Wai, Elizabeth I. Alger, Kevin A. Bird, Alan E. Yocca, Shujun Ou & Ning Jiang, Michigan State University; Thomas J. Poorten, Michael A. Hardigan, Nathan Pumplin, Charlotte B. Acharya, Glenn S. Cole & Steven J. Knapp**, University of California, Davis; Michael R. McKain, University of Alabama; Ronald D. Smith, Scott J. Teresi & Joshua R. Puzey, University of Arizona; Gil Ben-Zvi, Avital Brodt & Kobi Baruch, NRGene, Israel; Thomas Swale & Lily Shiue, Dovetail Genomics, California; Jeffrey P. Mower, University of Nebraska; Kevin L. Childs, Michigan State University; and Michael Freeling, University of California, Berkeley
** Corresponding authors
Image of Fragaria x ananassa Duchesne ex Rozier by Alpsdake, from the Wikimedia Commons under the Creative Commons Attribution-Share Alike 3.0 Unported license.

Leave a comment