By Kevin E. Noonan —
The orange clownfish, Amphiprion percula, is an important denizen of many reef systems (and, thanks to Disney, Pixar, and Ellen Degeneris, one of the most famous fishes since the Billy Bass). One of thirty species of anemonefishes in the family of damselfishes, the orange clownfish is found in northern Australia, including the Great Barrier Reef (GBR), and in Papua New Guinea, Solomon Islands, and Vanuatu. There is a related species, the false clownfish A. ocellaris, that occurs in the Indo‐Malaysian region, from the Ryukyu Islands of Japan, throughout South‐East Asia and south to north‐western Australia (but not the GBR). The orange clownfish has a mutualistic relationship with sea anemones, in particular, Stichodactyla gigantea and Heteractis magnifica.
The clownfish genome was recently explicated in an article published in Molecular Ecology Resources, entitled "Finding Nemo's Genes: A chromosome‐scale reference assembly of the genome of the orange clownfish Amphiprion percula." The study, by a number of researchers* haling from King Abdullah University of Science and Technology, Saudi Arabia; Australian National University, Canberra; and James Cook University, Queensland. The work is preliminary, insofar as while it describes the fish genome and provides comparisons with other fishes, there is as yet no understanding of the genetic basis for a variety of ecological and behavioral peculiarities of these fish, including patterns and processes of social organization; sex change; mutualism; habitat selection; lifespan; predator–prey interactions; scale of larval dispersal and population connectivity in marine fishes. The fish is recognized for its importance as a model member of reef fauna, and is expected to contribute to an better understanding of ecological effects of environmental disturbances in marine ecosystems, climate change, and particularly ocean acidification.
As reported in the study, the haploid chromosome number was set as 24, which is consistent with the observed haploid chromosome number of the Amphiprioninae, as published for A. ocellaris (Arai et al., 1976), A. frenatus, (Molina & Galetti, 2004; Takai & Kosuga, 2007), A. clarkii (Arai & Inoue, 1976; Takai & Kosuga, 2007), A. perideraion (Supiwong et al., 2015), and A. polymnus (Tanomtong et al., 2012). 98% of the orange clownfish genome was mapped, comprising 908.8 Mbp. Identified in the report are 26,597 protein-coding genes. The genome sequences are provided in an integrated database, the Nemo Genome Database (www.nemogenome.org). These genomic sequences are available for comparison with genome assemblies of two anemonefish, A. frenatus (Marcionetti et al., 2018) and A. ocellaris (Tan et al., 2018).
Both genomic and mitochondrial DNA was sequenced from clownfish brain tissue, and mitochondrial DNA was reported to comprise 16,638 bp, 13 coding genes, 22 tRNA genes, 12S and 16S rRNA sequences, and a D-loop control region. A. percula h mitochondria shares 95.5% sequence similarity to mitochondrial genomes with A. ocellaris.

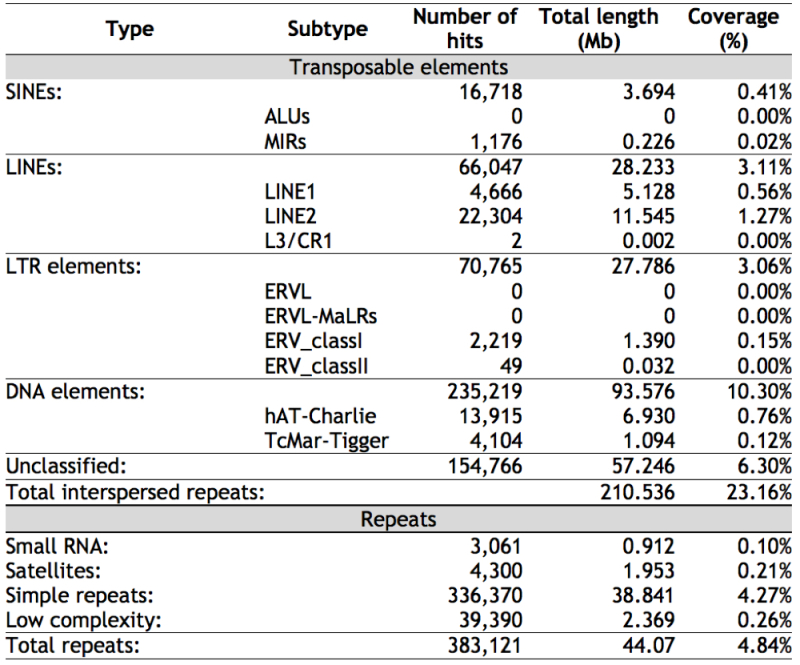
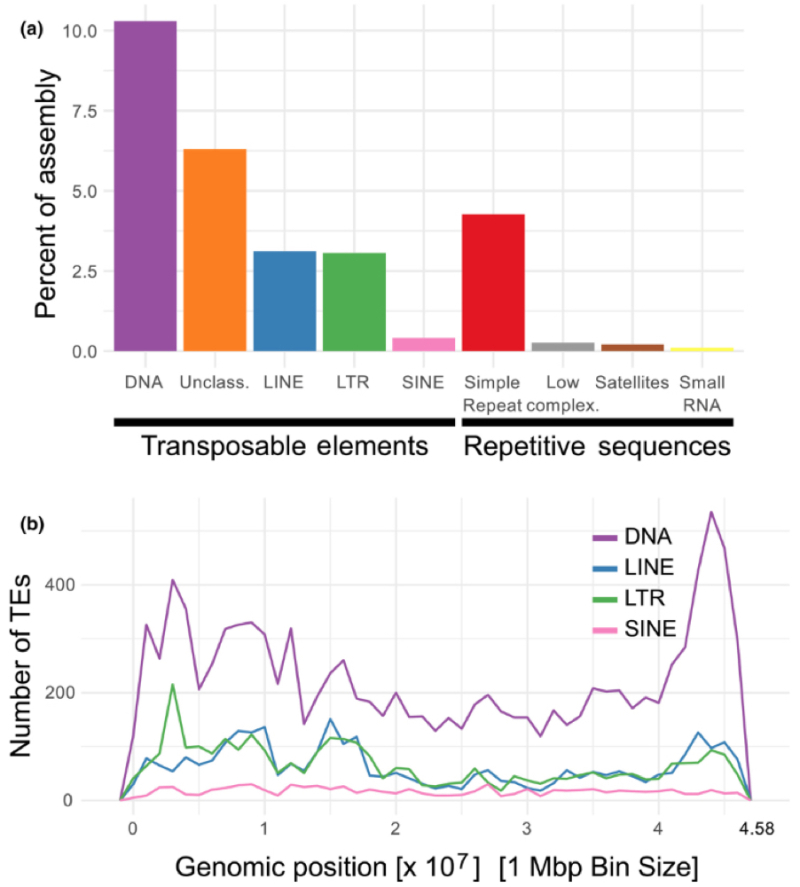
Clownfish genomic DNA was found to contain 21,644 repetitive sequences, making up 28% of the genome repetitive DNA, wherein 10% of these repeats derived from transposons (compared with ~ 3% in mammals). RNA sequence data was also obtained from 10 different tissues, showing 26,597 expressed genes from 35,478 transcripts. 8.1% of the clownfish genome comprises genome protein coding sequences, with a gene occurring on average every 30 Mbp.
Comparisons between clownfish genes and genes from four other species (Asian seabass, Nile tilapia, southern platyfish and zebrafish) showed that 89% of the genes could be assigned to 19,838 "orthologous" groups while 4,429 genes were specific for the orange clownfish. The largest gene identified in these studies was extracellular matrix protein FRAS1 (26.5 kb) and Titin was the largest encoded protein (18,851 amino acids). The authors acknowledge the preliminary nature of these results by stating "future investigations will focus on the characterization of these unique genes and what roles they may play in orange clownfish phenotypic traits."
*Robert Lehmann, Damien J. Lightfoot, Celia Schunter, Craig T. Michell,Hajime Ohyanagi, Katsuhiko Mineta Sylvain Foret, Michael L. Berumen,David J. Miller, Manuel Aranda,, Takashi Gojobori, Philip L. Munday, and Timothy Ravasi

Leave a comment